gRNA validation for wheat genome editing with the CRISPR-Cas9 system | BMC Biotechnology | Full Text

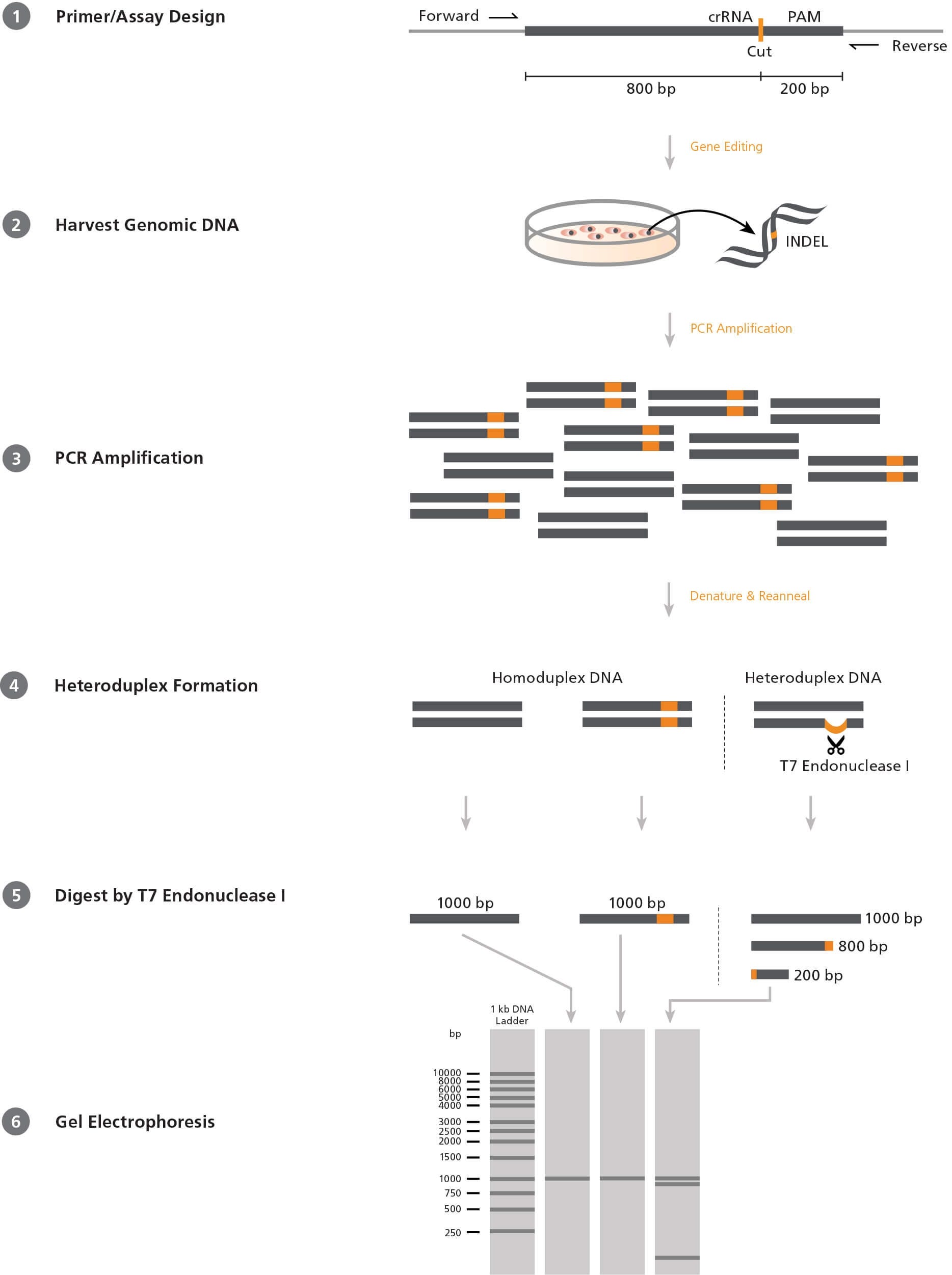
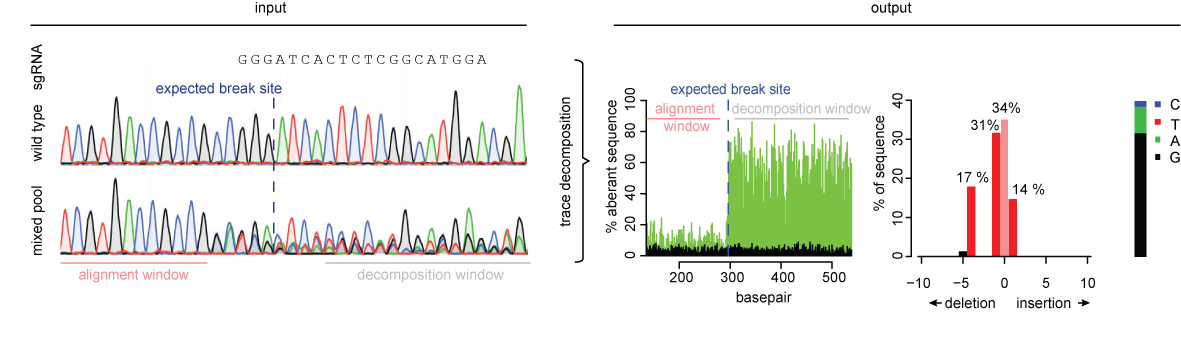
CRISRP-Cas9 activity reported with the T7E1 Assay and Next-Generation... | Download Scientific Diagram
Chromatin accessibility and guide sequence secondary structure affect CRISPR‐Cas9 gene editing efficiency
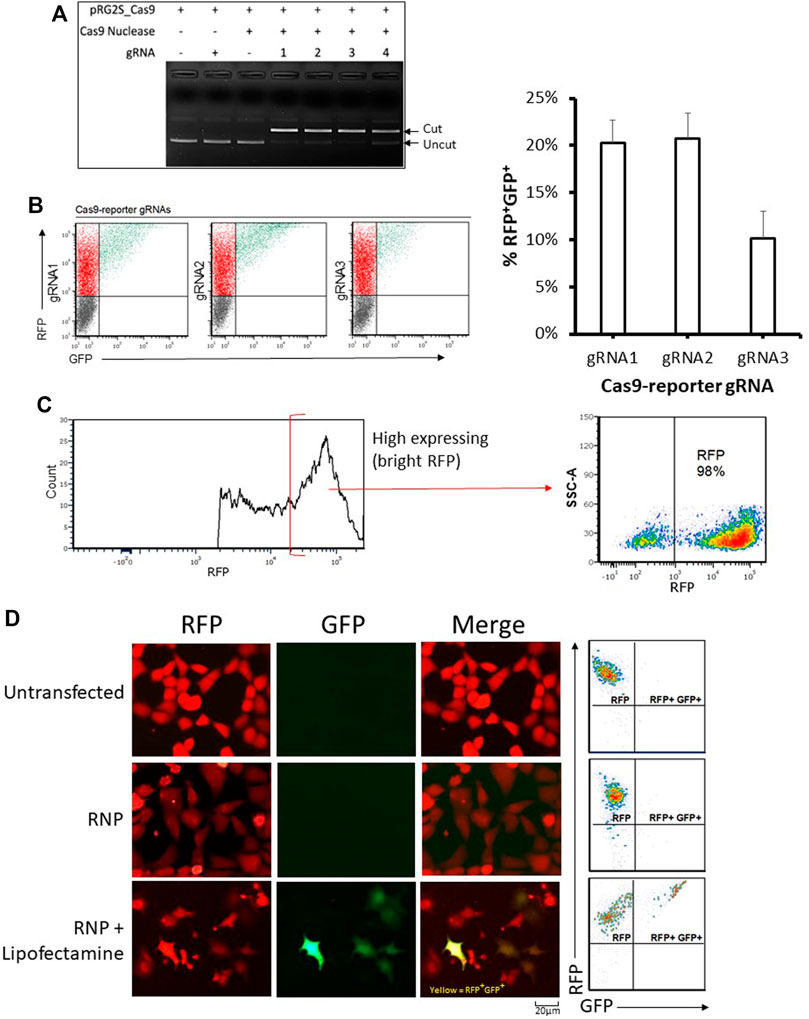
Frontiers | Rapid Assessment of CRISPR Transfection Efficiency and Enrichment of CRISPR Induced Mutations Using a Dual-Fluorescent Stable Reporter System | Genome Editing

A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020: Molecular Therapy - Nucleic Acids
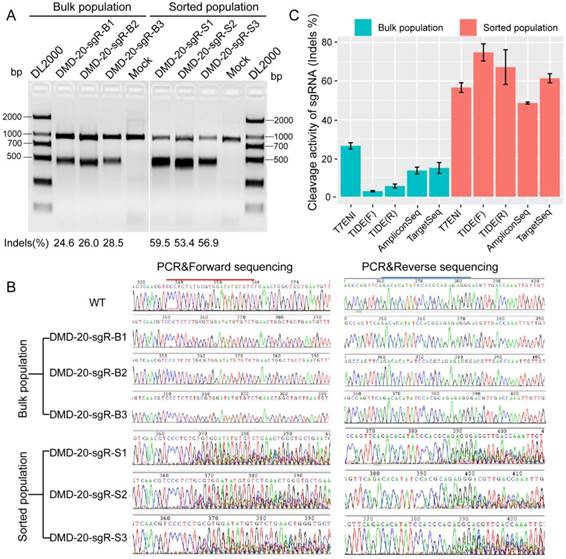
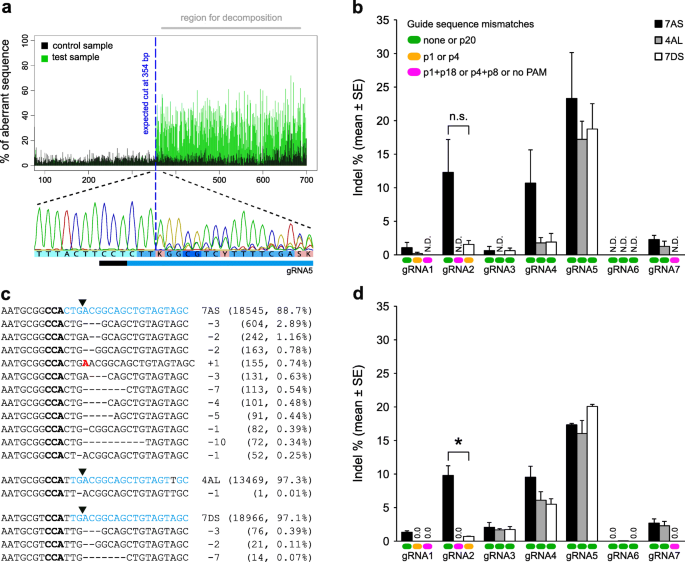
Evaluation of the effects of sequence length and microsatellite instability on single-guide RNA activity and specificity
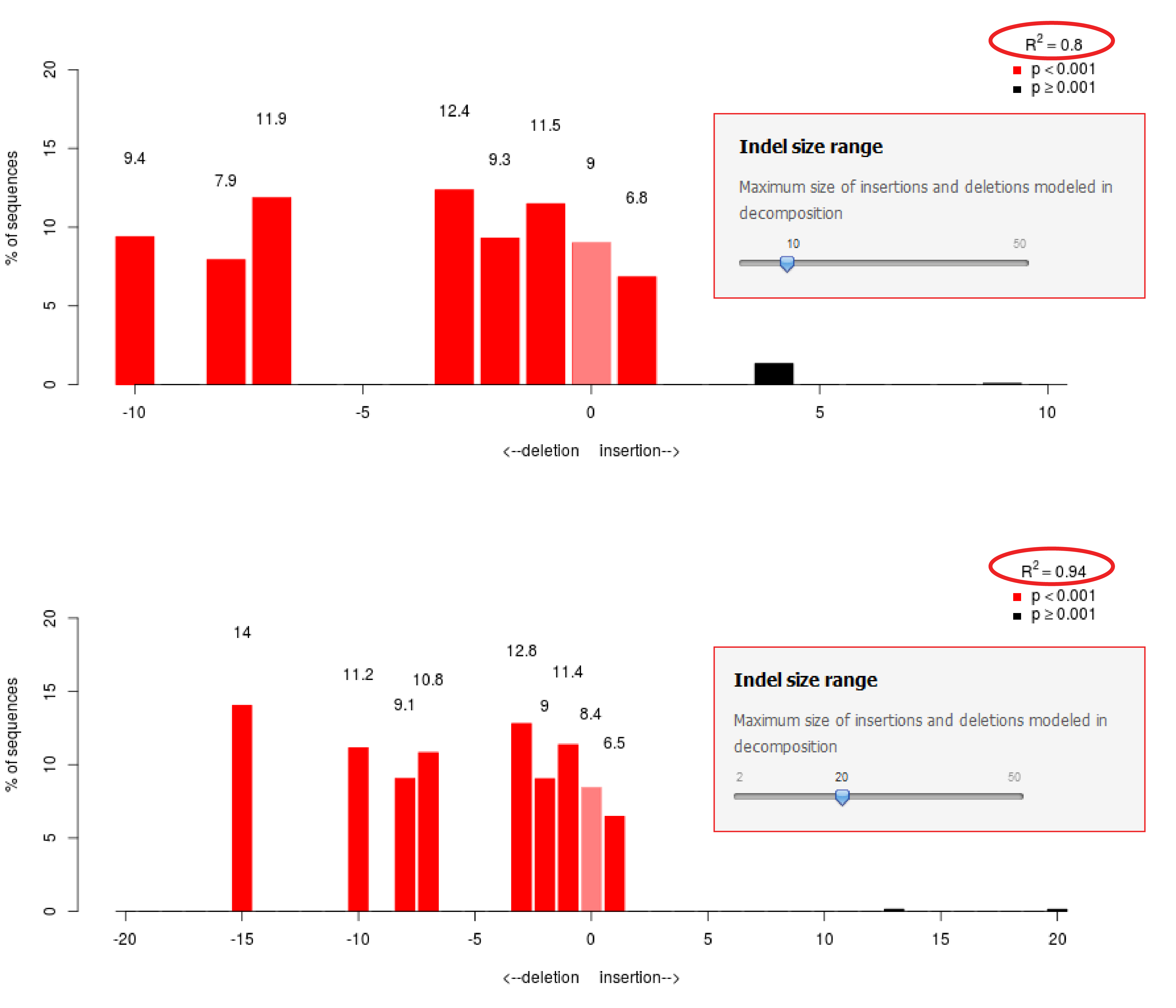
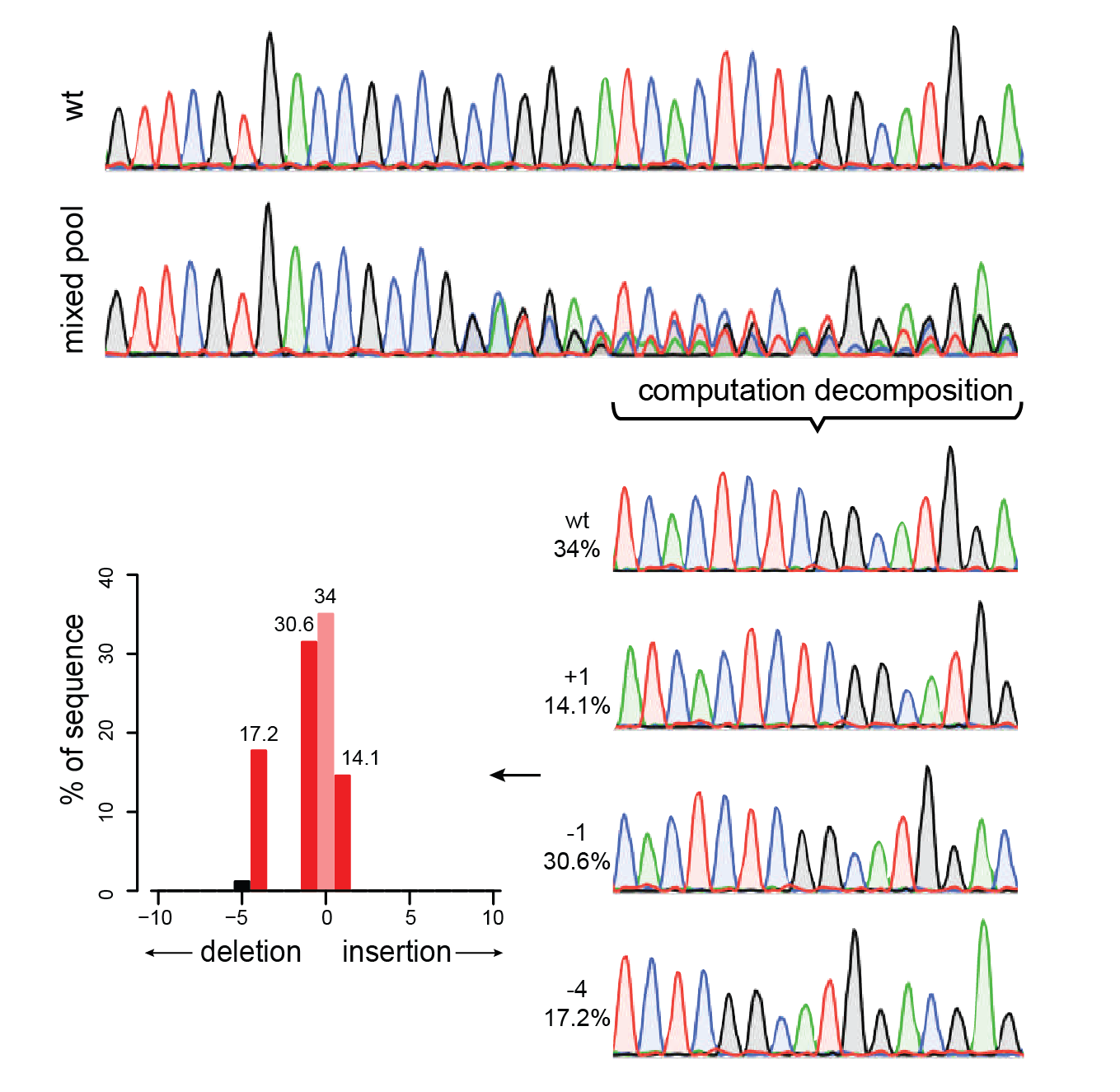
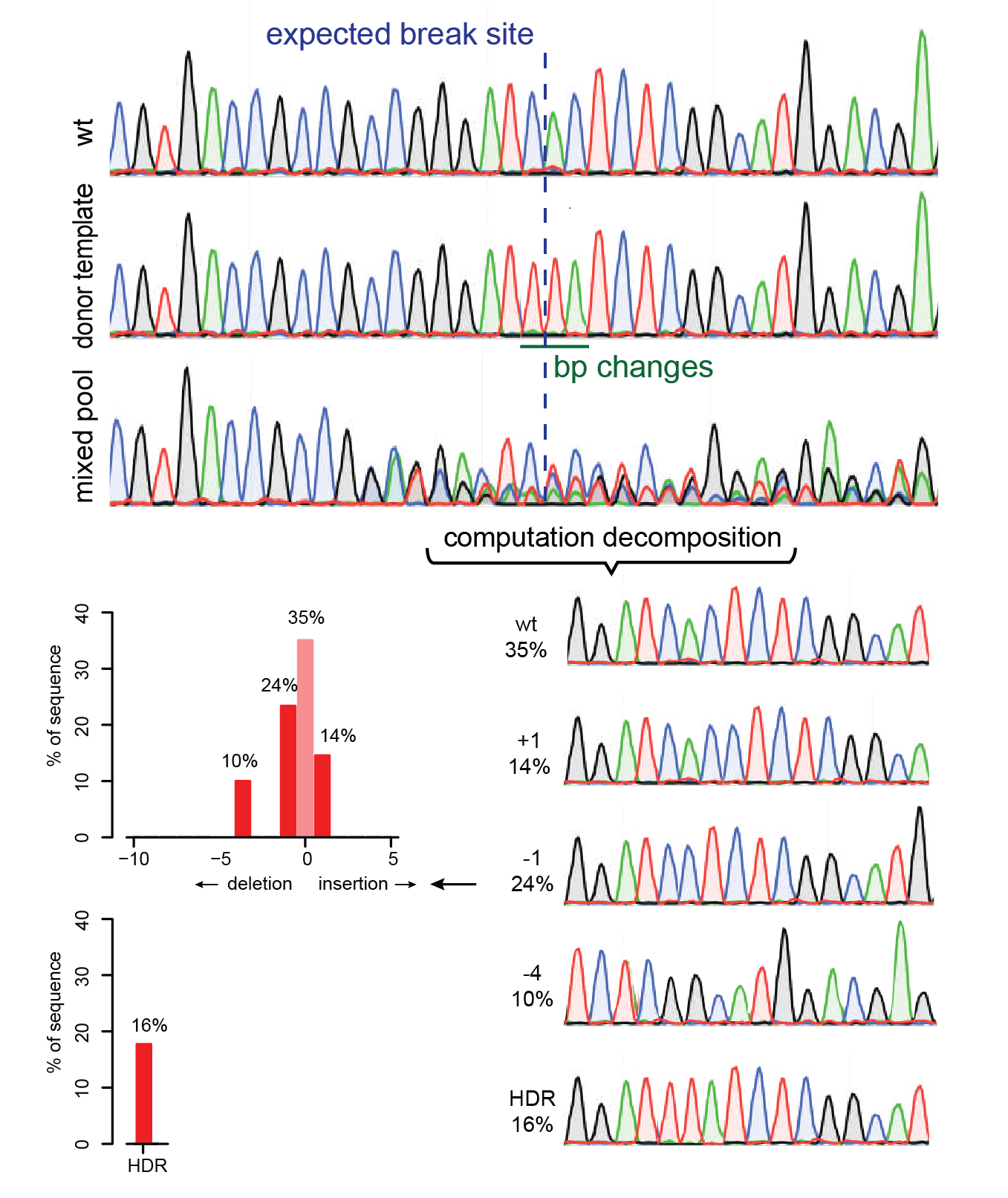
TIDE (tracking of indels by decomposition) analyses showing the indel... | Download Scientific Diagram

A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020: Molecular Therapy - Nucleic Acids
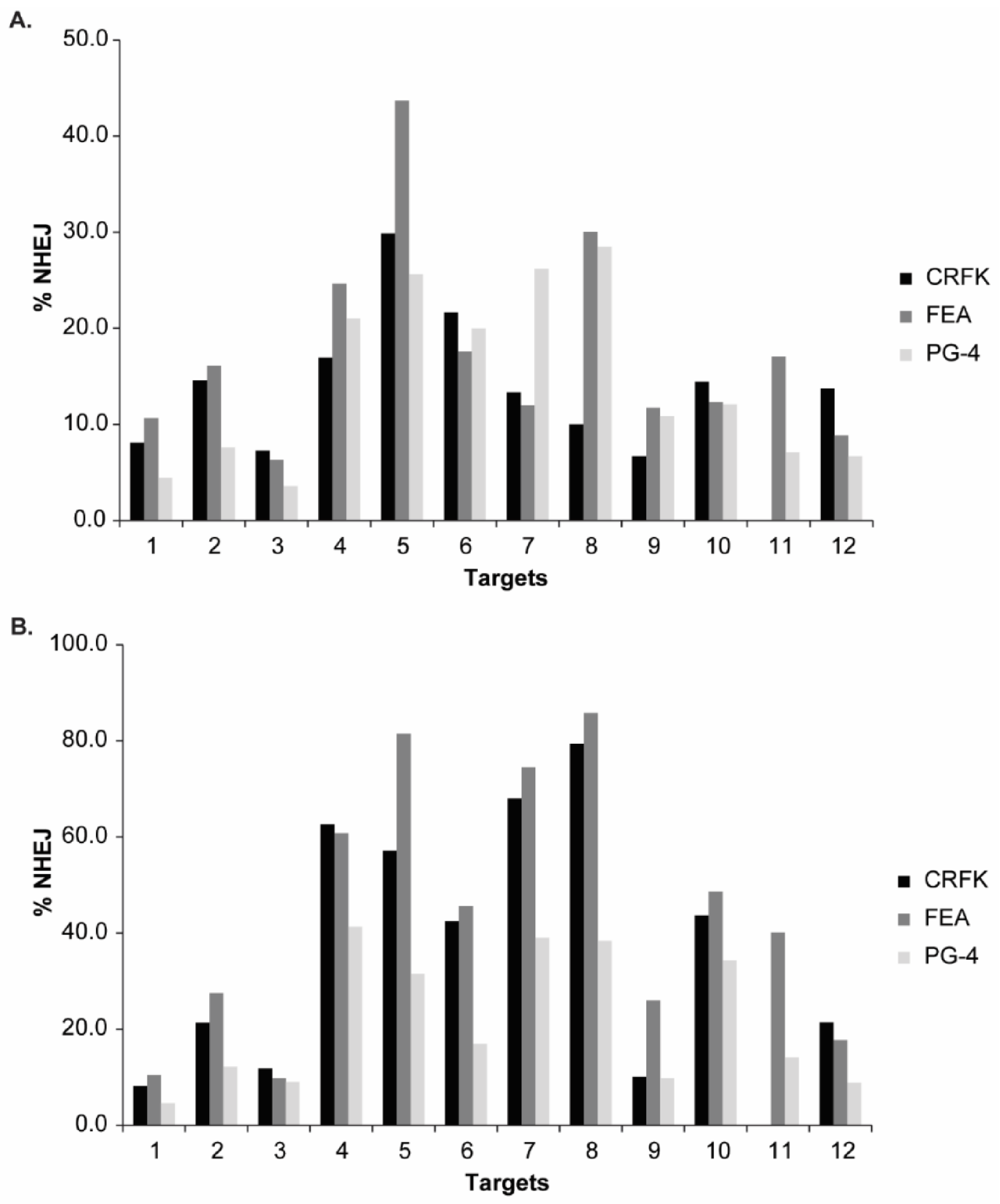
Viruses | Free Full-Text | Adeno-Associated Vector-Delivered CRISPR/SaCas9 System Reduces Feline Leukemia Virus Production In Vitro | HTML